For our international customers, please be advised that orders cannot be placed through our website by customers in countries with International Distributor representation.
Deoxyribonuclease I - Manual
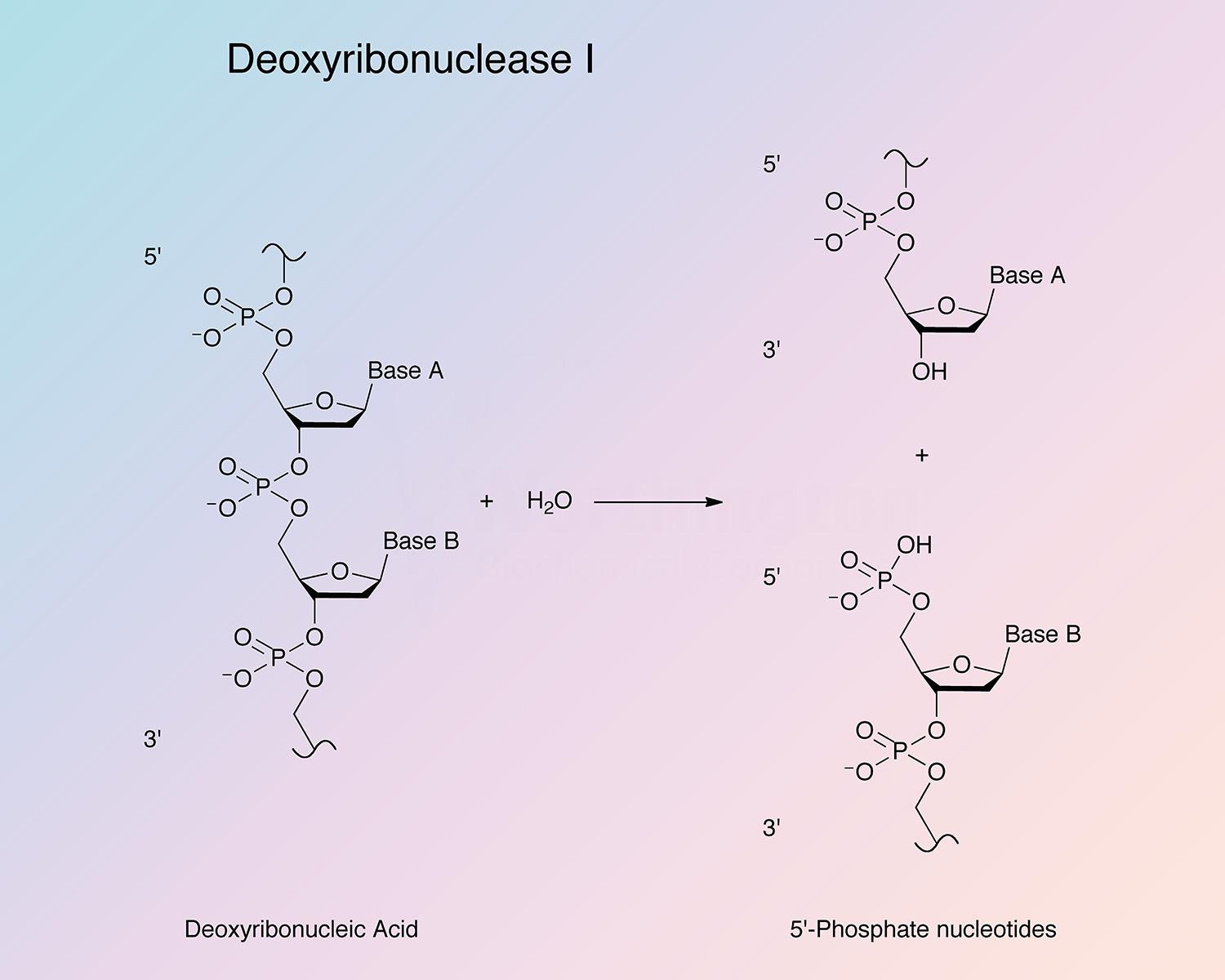
Bovine pancreatic deoxyribonuclease is an endonuclease that preferentially splits phosphodiester linkages adjacent to a pyrimidine nucleotide, yielding 5’-phosphate terminated polynucleotides with a free hydroxyl group at the 3’ position. DNase I is secreted by exocrine glands, and found most abundantly in the pancreas and parotid. It is also present in lower quantities in other tissues (Chen and Liao 2006, and Nadano et al. 1993).
DNase is known to be involved in apoptosis and has been proposed to play a role in the regulation of actin polymerization in cells. In addition to its use in molecular biology, DNase I has been used as a treatment for cystic fibrosis, and systemic lupus erythematosus (Chen and Liao 2006).
In 1903, Araki liquefied gels of alpha-nucleic acid with extracts of liver, spleen, and thymus tissue (Araki 1903, and Kunitz 1950). Plenge also showed similar activities in microorganisms (Plenge 1903). In 1913, De la Blanchardière developed an assay for the liquification of thymus DNA gel, and showed it was not accompanied by the destruction of DNA bases (Kunitz 1950).
Over the next few decades various authors described enzymes that cleaved nucleotides with names such as nucleinase (Levene and Medigreceanu 1911), nucleogelase (Feulgen 1923), polynucleotidase (Levene and Dillon 1932), desoxyribonucleodepolymerase (Greenstein 1943), thymonucleodepolymerase (Laskowski 1946), and desoxyribonuclease (McCarty 1946). In 1950, Kunitz first crystallized and coined today’s name, deoxyribonuclease.
In the early 1970s, Salnikow et al. found DNase I was present in four isoforms distinguishable by their sialic acid contents (Salnikow et al. 1970). Soon after, the polypeptide chain structure was elucidated (Salnikow et al. 1973b, Liao et al. 1973, and Catley 1973). The crystal structure was determined at a 2.0 Å resolution by Suck and Oefner in 1986 and later refined (Oefner and Suck 1986).
In 1990, the gene coding for Bovine pancreas DNase I (bpDNase I) was constructed from synthetic oligonucleotides, and then cloned and expressed in E. coli (Worrall and Connolly 1990). In 1992, the X-ray structure of the bpDNaseI-d(GGTATACC)2 complex was obtained at a 2.3 Å resolution (Weston et al. 1992). In 1998, the bpDNaseI gene was cloned from bp-cDNA and expressed in E. coli (Chen et al. 1998).
Today, researchers continue to investigate the residues involved in substrate binding and catalytic activity (Chen et al. 2008), novel inhibitors (Chen and Liao 2008), the specificity of DNAse I for specific DNA sequences (Heddi et al. 2010), and the conformational changes that occur in both DNAse I and DNA upon protein-DNA complexation (N’soukpoé-Kossi et al. 2008).
bpDNase I is not base nor sequence specific; however, it does not cleave randomly. It shows preference for cleavage at the 5’ side of pyrimidines, and is particularly pronounced in alternative copolymers (Bernardi et al. 1975, and Lomonossoff et al. 1981). It has been shown that variations in the twist angle are recognized by DNase I (Dickerson and Drew 1981). The specificity of DNase I also depends on the divalent cations present. In the presence of Ca2+ and Mg2+, it causes single strand breaks, and in the presence of Mn2+ double strand breaks have been reported (Junowicz and Spencer 1973, and Campbell and Jackson 1980).
bpDNase I exists as many isoforms, which differ both genetically and in sialic acid content (Liao 1974, Liao 1981, and Chang et al. 1994). The polypeptide chain is glycosylated at Asn-18 and because of the sialic acids of the carbohydrate side chain, shows charge heterogeneity (Chen and Liao 2006).
The enzyme is particularly stable due to an extended hydrophobic core, which is composed of two tightly-packed six-stranded beta-pleated sheets. These are surrounded by eight helices and several loop regions, which are stabilized by bound calcium atoms. The enzyme is further stabilized by intramolecular hydrogen bonds, salt bridges, and two disulfide bonds (Chen and Liao 2006).
Similar to human, mouse, and rat, the bovine pancreas DNase I gene consists of nine exons, and only the last eight encode the protein (De María and Arruti 2003). The nascent protein is directed to the secretory pathway organelles by a 22 amino acid signal sequence, encoded by exon two. The primary active site residue, H166, is encoded by exon six (De María and Arruti 2003, and Kraehenbuhl et al. 1977). Although exon lengths are almost equivalent in bovine and other mammalian species, the intron lengths vary greatly. A TATA box sequence is located 35 bp upstream of exon I (De María and Arruti 2003). It has been proposed that multiple splicing events that affect the coding sequence may be a mechanism to downregulate DNase I expression (Liu et al. 1997).
- DNA removal in primary cell isolation: decreases viscosity providing better yields
- DNA removal in bioprocessing applications
- Removing genomic DNA from RNA preparations prior to RT-PCR
- in vitro transcription
- Nick translation
- DNase footprinting
- Actin binding
- UV crosslinking of proteins to nucleic acids
- Radioactive labeling
P00639
- Class: Alpha Beta
- Architecture: 4-Layer Sandwich
- Topology: Deoxyribonuclease I; Chain A
29.1 kDa (Theoretical)
7.8
5.08 (Theoretical)
- 36,750 cm-1 M-1
- E1%,280 = 11.1
- Tyrosine (Y87)
- Glutamate (E100)
- Histidine (H156)
- Bivalent metal ions (Junowicz and Spencer 1973b, Poulos and Price 1972, Price 1969, and Price 1975)
- 2-mercaptoethanol (Laskowski 1971)
- 2-nitro-5-thiocyanobenzoic acid (Moore 1981)
- Actin (Mannherz et al. 2008, and Lazarides and Lindberg 1974)
- Alfatoxin B2a, G2, G2a, and M1 (non-competitive) (Schabort 1970)
- EGTA (Moore 1981) and EDTA (Junowicz and Spencer 1973)
- Sodium dodecyl sulfate (Liao 1975a)
- Calf spleen inhibitor protein (Laskowski 1971)
- Carbodiimide and cholesterol sulfate (Iwamori 2000)
- Iodoacetate (Moore 1981)