For our international customers, please be advised that orders cannot be placed through our website by customers in countries with International Distributor representation.
Amino Acid Oxidase, D- - Manual
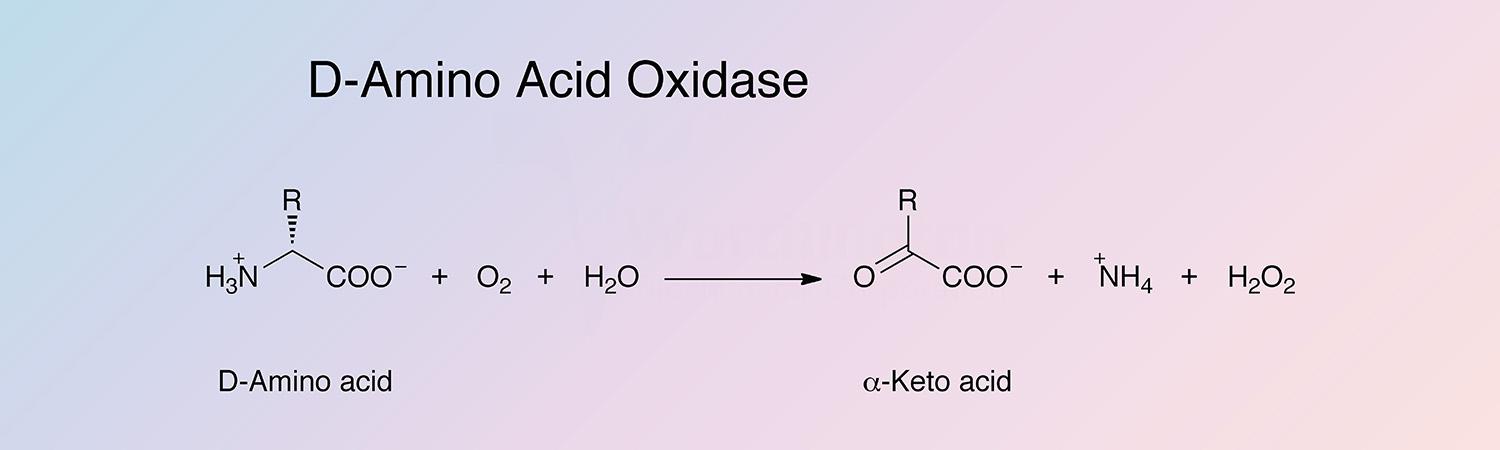
D-amino acid oxidase (DAAO) is an oxidoreductase that oxidatively deaminates D-amino acids to the corresponding alpha-keto acid. DAAO is a flavoenzyme, containing 1 mole of FAD per monomer.
D-amino acid oxidase was the second flavoenzyme to be discovered. In 1935, Krebs discovered DAAO after he observed that porcine kidney homogenates deaminated amino acids of the “d-series... much more rapidly than the natural isomerides.” In 1938, Warburg and Christian identified FAD as the enzyme's cofactor.
In the 1940s, Singer tested the role of sulfhydryl reagents as inhibitors (Singer 1948). In 1958, Kubo et al. developed a purification procedure for pkDAAO (porcine kidney D-amino acid oxidase). In 1961, Massey et al. improved this method, and it was improved again by Curti et al. (Curti et al. 1973). Curti et al. also first reported 5 sulfhydryl groups and 8 tryptophans per mole FAD. Tu and McCormick indicated that a tyrosine residue was probably involved in the active site, and Yagi et al. reported the monomer to be more active than the dimer (Tu and McCormick 1973, and Yagi et al. 1973b).
In the early 1980s, various inhibitors were investigated (Alston et al. 1983, and Carrea et al. 1983), and the amino acid sequence was preliminarily identified (Ronchi et al. 1982). Beginning in the mid-1980s, studies on DAAOs of microorganisms made it possible to carry out detailed biochemical studies (Pilone 2000). In the late 1980s, DAAO was expressed in E. coli by Ciccarelli et al., and molecular cloning and sequence analysis of cDNAs were performed by Fukui et al. (Fukui et al. 1987, and Ciccarelli et al. 1989).
In 1996, two groups independently reported on the 3D structure of pkDAAO, and Mettevi et al. provided strong support for a hydride mechanism (Mattevi et al. 1996, and Mizutani et al. 1996). Site directed mutagenesis studies (Harris et al. 1999, Molla et al. 2000, and Boselli et al. 2002) and the high resolution structure of yeast Rhodotorula gracilis support this hypothesis (Umhau et al. 2000). In 2006, Kawazoe et al. published the 3D structure of human DAAO.
Current research on DAAO is focused on its use as a biocatalyst (Truppo et al. 2009) and as a component of biosensors (van Staden et al. 2010, and Inaba et al. 2003).
The D-isomers of proline, methionine, isoleucine, alanine, valine and phenylalanine are good substrates (Scannone et al. 1964, and Dixon and Kleppe 1965b). The enzyme is reported to act on L-proline (Wellner and Scannone 1964) and D-lactate (Yagi and Ozawa 1964b). The best substrate for pkDAOO is D-proline, and DAAOs exhibit very poor or no activity toward D-aspartate (Tishkov and Khoronenkova 2005).
The substrate-binding domains in various species’ primary structures do not show high homology. This may reflect the wide variation in specificities observed for DAAOs from different origins (Tishkov and Khoronenkova 2005).
The active pkDAAO holoenzyme is a monomer of 347 amino acids that can undergo dimerization. The monomer has been found to be more active than the dimer, and contains 1 mol FAD noncovalently bound per monomer. All DAAOs characterized as of 2000 contain noncovalently bound FAD as their prosthetic group (Pilone 2000).
The gene encoding mammalian DAAO is present in a single copy in the genome. A 1041 bp open reading frame encodes all 347 amino acids of the enzyme. This indicates posttranslational processing by proteolytic enzymes does not occur (Fukui 1987).
The primary structure of porcine D-amino acid oxidase was determined by Ronchi et al. (Ronchi et al. 1982), and the gene was cloned by Momoi et al. (Momoi et al. 1988). There are six regions of the primary structure that are highly conserved in DAAOs of various sources (Faotto et al. 1995). Regions I contains the consensus sequence GXGXXG, and both regions I and III have been found to be involved in coenzyme binding (Wierenga et al. 1983). Regions II, IV, and V contain the active site residues. The Ser-Lys/His-Leu terminal sequence is the peroxisomal targeting signal sequence (Subramani 1993, and Pilone 2000)
Mammalian DAAOs show 63% identity, and the three known DAAOs of microorganisms (R. gracilis, T. variabilis, and Fusarium solanii) show a 18% identity. 30% identity is observed between yeast and mammalian DAAOs (Pilone 2000).
- Keto acid preparation
- Oxidation reduction studies
- Separation of L-amino acids from racemic mixtures
- FAD determination
- D-alanine determination
- Biosensors (Inaba et al. 2003)
P00371
- Class: Alpha Beta
- Architecture: 2-Layer Sandwich and 3-Layer (aba) Sandwich
- Topology: D-Amino Acid Oxidase; Chain A, domain 2 and Rossmann fold
- 78.7 kDa (Theoretical)
- Monomeric: 38.0-39.0 kDa (Curti et al. 1973, and Tu et al. 1973)
Dependent on the substrate: approximately 9 for D-alanine (Dixon and Kleppe 1965c).
7.0, 7.2 (Tishkov and Khoronenkova 2005)
- 75,420 cm-1 M-1 (Theoretical)
- E1%,280 = 19.17 (Theoretical)
- Tyrosine (Y224)
- Aspartic acid (D228)
- Arginine (R283)
(Pilone 2000)
- 2-hydroxy acids, 2-oxo acids, and 2-oxobutyrate (Dixon 1965b)
- Metabolites and drugs (Hamilton and Buckthal 1982)
- Adenosine 5’-monophosphate and aniline (Yagi et al. 1972c)
- Benzoate (Pollegioni et al. 2007)
- Sodium benzoate (Nguyen et al. 2009)