For our international customers, please be advised that orders cannot be placed through our website by customers in countries with International Distributor representation.
Papain, Chymo - Manual
Chymopapain is an extracellular cysteine proteinase secreted in the latex of the Carica papaya tree. Chymopapain was thought to exist as two forms, A and B (Maes et al. 1996); however, those terminologies were replaced through the results of molecular cloning experiments, which revealed a multiplicity of enzymes differing in only one or two amino acid residues (Taylor et al. 1999).
In 1941, Jansen and Balls first described chymopapain, after obtaining it in crystalline form from papaya latex (Guha et al. 2006). It was termed chymopapain to distinguish it as the proteolytic activity remaining after the removal of papain (Taylor et al. 1999).
In 1967, Kunimitsu and Yasunobo first demonstrated chymopapain’s charge heterogeneity through cation-exchange chromatography, creating the notion that two forms of the enzyme existed: chymopapain A and chymopapain B. It has since been determined the separation observed during cation-exchange chromatography was instead due to the charge heterogeneity of chymopapain, and contamination with glycyl endopeptidase (Buttle 2004).
The amino acid sequences for chymopapain were published by Jacquet et al. in 1989 (Jacquet et al. 1989b) and Watson et al. in 1990. In 1996, Maes et al. elucidated the structure of chymopapain to a 1.7 Å resolution, and in 1999 Taylor et al.’s molecular cloning studies revealed five isoforms of the enzyme, putting an end to the chymopapain A and chymopapain B terminologies.
Recent research continues to investigate the wound-induced expression of chymopapain, as well as other papaya endopeptidases (Azarkan 2006). Chymopapain is also being used as a control to study human invertebral disc degeneration, as it mimics the natural process of degradation (Chen et al. 2009). Chymopapain has also found use in research in rheumatoid arthritis as it mimics the increased activity of proteases found in the joints of patients with rheumatoid arthritis (Sabaratnam et al. 2007).
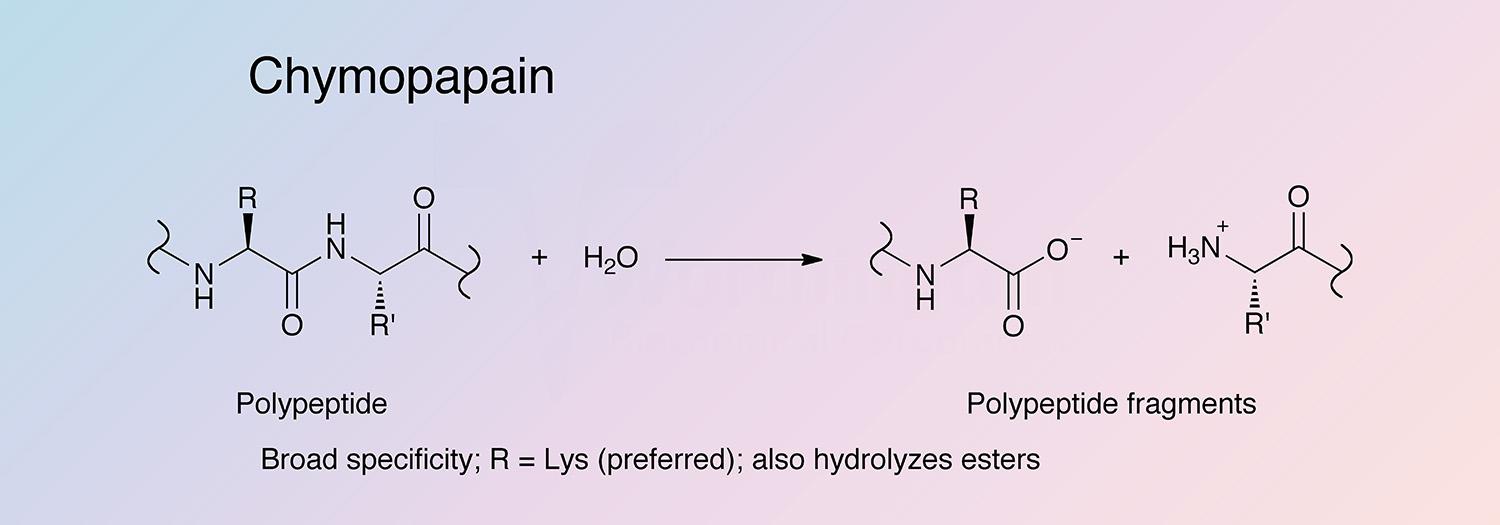
Chymopapain shows a broad substrate specificity (Jacquet et al. 1989a). Chymopapain hydrolyzes a wide variety of substrates, similar to papain, but at slower rates. The substrate specificity is primarily controlled by the S2 subsite (Drenth et al. 1976).
Chymopapain is synthesized with N-terminal signal and propeptide regions, which are required for proper folding (McKee et al. 1986, Revell et al. 1993, Karrer et al. 1993, Taylor et al. 1995, and Vernet et al. 1995). These propeptides have been found to be selective high-affinity inhibitors of the mature enzyme (Taylor et al. 1995). Unlike other proteinases that have one free cysteine in the active site, chymopapain has a second non-essential free cysteine residue (C117) (Sumner et al. 1993).
The enzyme is a polypeptide chain of 218 amino acids. Chymopapain shows 58% sequence identity with papain (Sumner et al. 1993, and Buttle 2004). Among the papaya proteinases, V133 and V157 are highly conserved. Chymopapain is unique in that V133 and V157 are replaced by leucine in all five isoforms (Taylor et al. 1999, and Buttle 2004).
- Rheumatoid arthritis research
- Control in disc degeneration studies
- Prior to being discontinued in the United States in 2003, chymopapain was injected directly into herniated disks to dissolve part of the disc and relieve pain
P14080
- Class: Alpha Beta
- Architecture: Alpha-Beta Complex
- Topology: Cathepsin B; Chain A
- 27 kDa (Ebata 1962)
- Broad, centered around 7 (Buttle 2004)
- 10.2-10.6 (Baines and Brocklehurst 1982)
- 42,600 cm-1 M-1 (Robinson 1975)
- E1%, 280 = 18.14 (Theoretical)
- Cysteine (C159)
- Histidine (H293)
- Asparagine (N313)
- 2,3-Dimercaptopropanol (Ebata and Yasunobu 1962)
- Cysteine (Ebata and Yasunobu 1962)
- Glutathione (Ebata and Yasunobu 1962)
- NaN3 (Kunimitsu 1970)
- Ag+, Cu2+, Hg2+, and Zn2+ (Kunimitsu 1970)
- Cystatin (Björk and Ylinenjörvi 1990)
- Iodoacetate
- PCMB
- Albumin and succinyl-albumin (Zucker et al. 1985)