For our international customers, please be advised that orders cannot be placed through our website by customers in countries with International Distributor representation.
Elastase - Manual
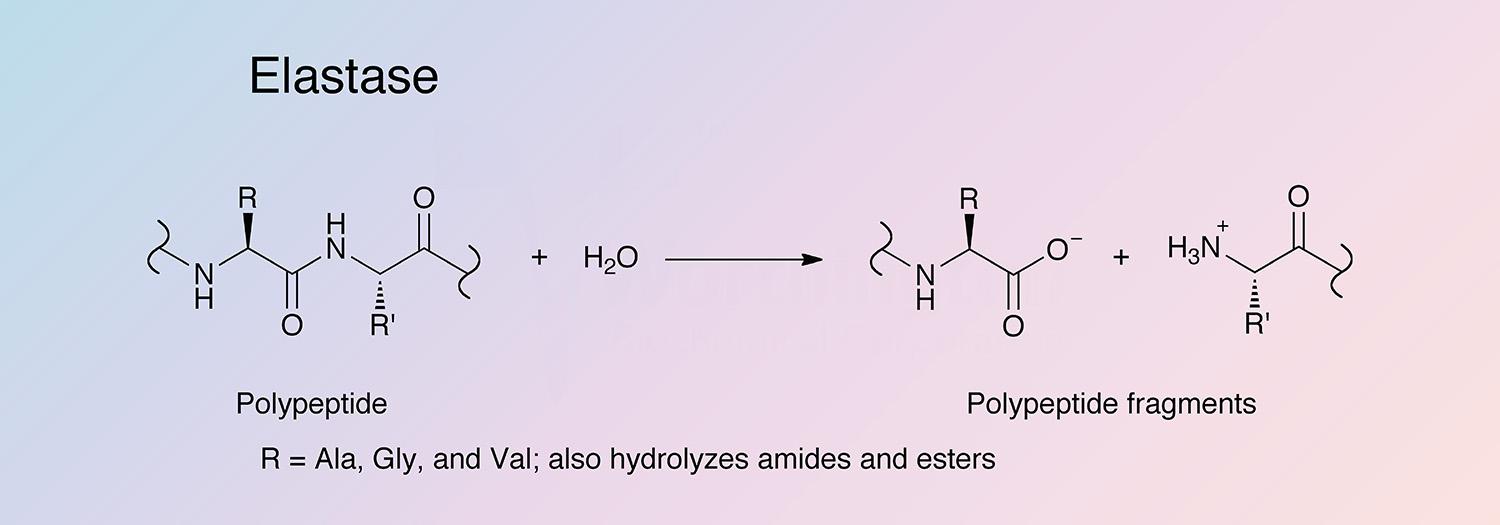
Elastase is a serine protease that also hydrolyzes amides and esters. It is produced in the pancreas as an inactive zymogen, and activated in the duodenum by trypsin. The following information applies to porcine elastase.
Eijkman is believed to be the first to have studied elastase as a product of bacteria (Eijkman 1904). However, it was not shown to differ from other proteolytic components of the pancreas, such as trypsin and chymotrypsin, until the Baló and Banga publication in 1949.
In 1968, Shotton and Hartley were the first to obtain crystalline porcine elastase. In the late 1960s and into the early 1970s, elastase’s involvement in emphysema, atherosclerosis, and acute hemorrhagic pancreatitis was investigated (Janoff and Sherer 1968, Janoff and Basch 1971, Talamo et al. 1971, Kaplan et al. 1973, and Bieth et al. 1974). In 1974, Ardelt discovered a second elastolytic proteinase, naming it pancreatic “elastase II” and the original “elastase I”.
In the 1980s, the catalytic properties of elastase were studied (Ascenzi et al. 1983), and inhibitors were identified (Largman et al. 1980, Lestienne et al. 1981, Kettner and Shenvi 1984, Poncz et al. 1984, and Imperiali and Abeles 1986). Elastase I and II genetic information was studied into the 1990s (Ornitz et al. 1985, Stevenson et al. 1986, Swift et al. 1984, Tani et al. 1987, and Gestin et al. 1997).
Elastase has recently been used in investigations of the mechanical forces and enzyme activity in extracellular matrix breakdown (Jesudason et al. 2010). Its use in artificial organ development has also been studied (Tedder et al. 2010), and it is used as a model to investigate the catalytic activity of the serine proteases (Tamada et al. 2009).
Porcine elastase I is specific for Ala-Ala and Ala-Gly bonds, while elastase II has a broad specificity for substrates with medium to large hydrophobic amino acids in the P1 position (Gertler et al. 1977, Del Mar et al. 1980, and Gestin et al. 1997). Porcine elastase is the most potent elastase, having a rate 20-fold higher than that of human leukocyte elastase (Bieth 1978, Bieth 1986, and Largman 1983).
Hydrolysis occurs in several steps. An adsorption complex between elastase and its substrate is formed, followed by nucleophilic attack (S214) to form an acyl-enzyme intermediate, and release of the first product (the C-terminal end of the substrate). The intermediate is hydrolyzed in a deacylation step, regenerating the active enzyme and releasing the second product (Bieth 1986).
The catalytic triad is formed by three hydrogen-bonded amino acid residues (H71, D119, and S214). The polypeptide chain is composed of two antiparallel beta-barrel domains, which form a crevice containing the catalytic triad, and a small proportion of alpha-helices (Bieth 2004).
Porcine pancreatic elastase is composed of a single peptide chain of 240 amino acids, and contains 4 disulfide bridges (Sawyer et al. 1973). It has a high degree of sequence identity with pancreatic elastases from other species, such as rat with whom it shares 86% identity (MacDonald et al. 1982). Elastase I and II genes share sequence similarity, especially in the 5’ proximal flanking regions, which include the TATA box and a putative tissue-specific enhancer sequence (Ornitz et al. 1985, Stevenson et al. 1986, Swift et al. 1984, Tani et al. 1987, and Gestin et al. 1997).
- Tissue dissociation: Because elastin is found in highest concentrations in the elastic fibers of connective tissues, elastase is frequently used to dissociate tissues that contain extensive intercellular fiber networks. For this purpose, it is usually used with other enzymes such as collagenase, trypsin, and chymotrypsin.
- Membrane protein solubilization
- Protein sequence studies
P00772
- 26.0 kDa (Bieth 2004)
- 8.5
- 9.5 (Bieth 2004)
- 54,870 cm-1 M-1 (Theoretical)
- E1%, 280 = 21.18 (Theoretical)
- Histidine (H71)
- Aspartic acid (D119)
- Serine (S214)
- Competitively inhibited by derivatives of dipeptides of alanine, valine, leucine, and isoleucine
- Inhibitors from natural sources (e.g. serpins) (Tsunemi et al. 1996, Matern et al. 2003, and Dementiev et al. 2006)
- Modified natural inhibitors (Ay et al. 2003, and Hilpert et al. 2003)
- Sulfate-modified lipids (Ito et al. 1998)