For our international customers, please be advised that orders cannot be placed through our website by customers in countries with International Distributor representation.
Adenosine Deaminase - Manual
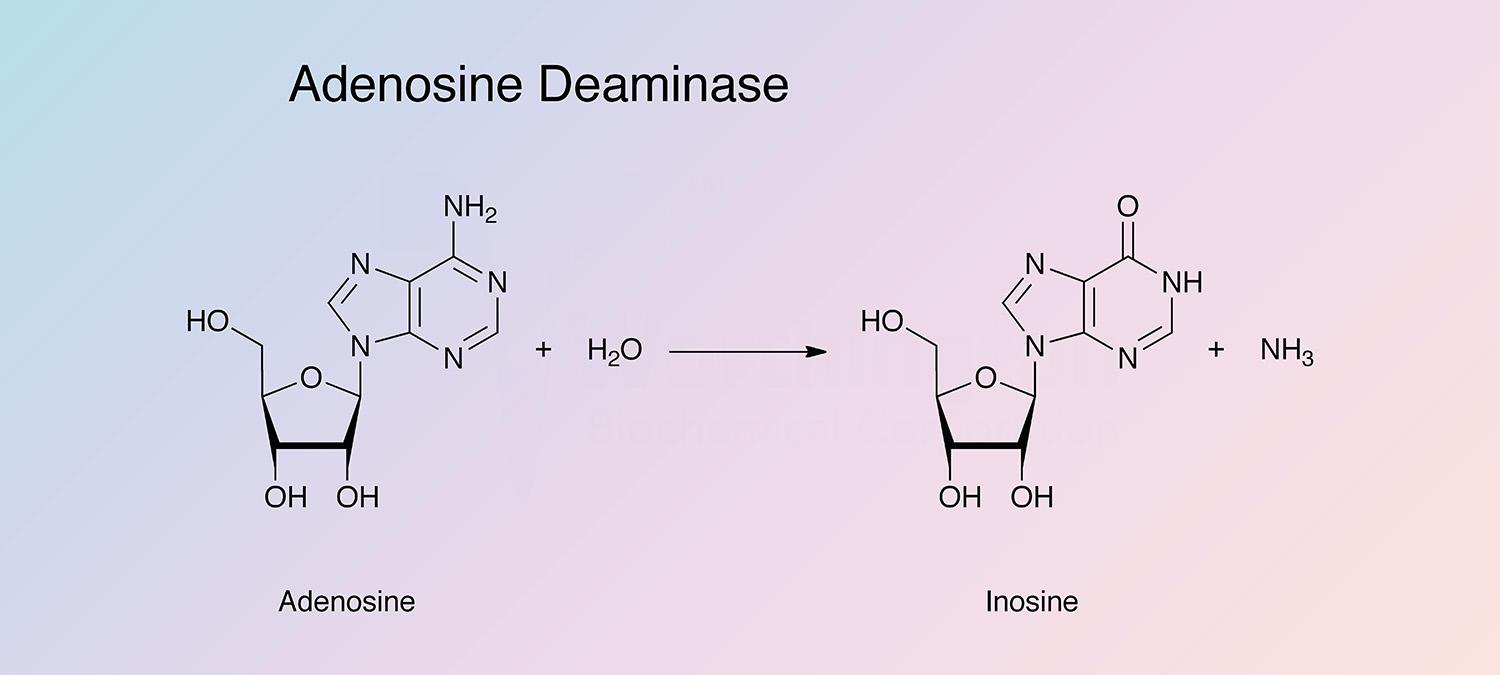
Adenosine deaminase catalyzes the irreversible deamination of 2'-deoxyadenosine and adenosine to deoxyinosine and inosine. It is found in a wide variety of prokaryotes and eukaryotes in different forms (Pospisilova and Frebort 2007).
In 1928 and in 1932, Schmidt first showed the high specificity of adenosine deaminase (Schmidt 1928 and 1932). Conway and Cook found that unlike other deaminases studied at the time, adenosine deaminase was not as affected by tissue inhibitors, phosphate, or bicarbonate (Conway and Cook 1939). It was later discovered that the tissue Schmidt had been using was a very poor source of adenosine deaminase, and Conway and Cook discovered the high activity in calf mucosa.
Throughout the 1940s and 1950s, the reaction mechanism was further investigated, and adenosine deaminases from different sources were purified and studied (Kaplan 1952 and 1955).
In the 1960s, the preparation of calf spleen adenosine deaminase was developed and refined, which led to a greater understanding of its enzymatic properties (Pfronger 1967 a and b). In the late 1960s, adenosine deaminases from different species were compared (Wolfendental 1968, Akedo et al. 1970, and Edwards et al. 1971), and into the 1970s research began to focus on adenosine deaminase from humans to better understand its connection to immunodeficiencies (Meuwissen and Pollara 1974, and Hirschhorn et al. 1976).
Research of the 1980s increased the use of adenosine deaminase as a diagnostic tool in a variety of illnesses including meningitis and tuberculosis (Ribera et al. 1987, and Segura et al. 1989). Inhibitors of adenosine deaminase were investigated in the 1990s (Weitberg and Corvese 1990), and in 1991 the crystal structure of mouse adenosine deaminase was resolved (Wilson et al. 1991).
Today, human adenosine deaminase is widely studied because of its importance in the medical field. Gene regulation and gene therapy are are being investigated to understand the clinical implications of ADA gene regulation (Aiuti et al. 2009, and Wang 2009).
Adenosine deaminase has evolved to act upon adenosine, and in the lab on its derivatives (Sharoyan et al. 2006). Additionally, adenosine deaminase acts on carbon-nitrogen bonds other than peptide bonds, in cyclic amidines (White and White 1997).
Adenosine deaminase has a (beta/alpha)8 barrel structural motif, and contains a zinc atom in the catalytic pocket. A single, bound, divalent cation (zinc or cobalt) is required for catalytic activity (Cristalli et al. 2001).
In higher eukaryotes, two different isozymes are encoded by different genes (Maier et al. 2005). In humans, ADA1 is a single-chain Zn-binding protein and almost all activity is attributed to this protein. ADA2 is believed to be produced by monocytes and is found in negligible quantities. Mutations in the ADA1 gene, where expression is blocked, cause immunodeficiency, whereas mutations that cause overexpression cause hemolytic anemia (Pospisilova and Frebort 2007).
The enzyme has a wide phylogenetic distribution and its sequence is highly conserved from bacteria to humans (Chang 1991). The genes from lower eukaryotes contain no introns, whereas 95% of the DNA sequence in mammalian ADA genes are occupied by introns (Pospisilova and Frebort 2007). Gene sequences have been reported for cattle, mouse, human, E. coli, Saccaromyces cerevisiae, and Streptomyces virginiae. The amino acid residues around the active site are highly conserved in mammals (Cristalli et al. 2001). Human and bovine adenosine deaminases are 93% identical (Kelly et al.1996), and human and mouse adenosine deaminases are 83% superimposible (Wiginton et al. 1984, Daddona et al. 1984, and Yeung et al. 1985). The bacterial enzyme shares 33% identity with mammalian adenosine deaminase (Kelly et al. 1996).
- Elevated levels are a diagnostic test for various human diseases (Ribera et al. 1987, and Segura et al. 1989)
- Inosine, deoxyinosine and dideoxyinosine synthesis (Sheid 1996)
- Determination of adenosine in biological fluids (Piras 1973)
- Determination of 5’-nucleotidase in blood (Blackburn et al. 1992)
P56658
- Class: Alpha Beta
- Architecture: Alpha-Beta Barrel
- Topology: TIM Barrel
- 40.8 kDa (Theoretical)
5.0-9.0 (Rossi and Lucacchini 1974)
- 5.33 (Theoretical)
43,960 cm-1 M-1
- Histidine (H214)
- Cysteine (C262)
- Aspartic acids (D295 and D296)
- Zn2+
- Ag+, Hg2+, Cu2+
- TPCK, DFP
- Sulfhydryl reagents
- Erythro-9-(2-hydroxy-3-nonyl)adenine and Erythro-9-(2-hydroxy-3-nonyl)-3-deazaadenine
- 9-(p-aminobenzyle)adenine and p-chloromercuribenzoate
- Purine riboside